Table of Contents (click to expand)
‘Junk’ DNA refers to non-coding DNA segments. It is actually a misnomer, as this ‘junk’ DNA has been found to play an important role in regulating gene expression.
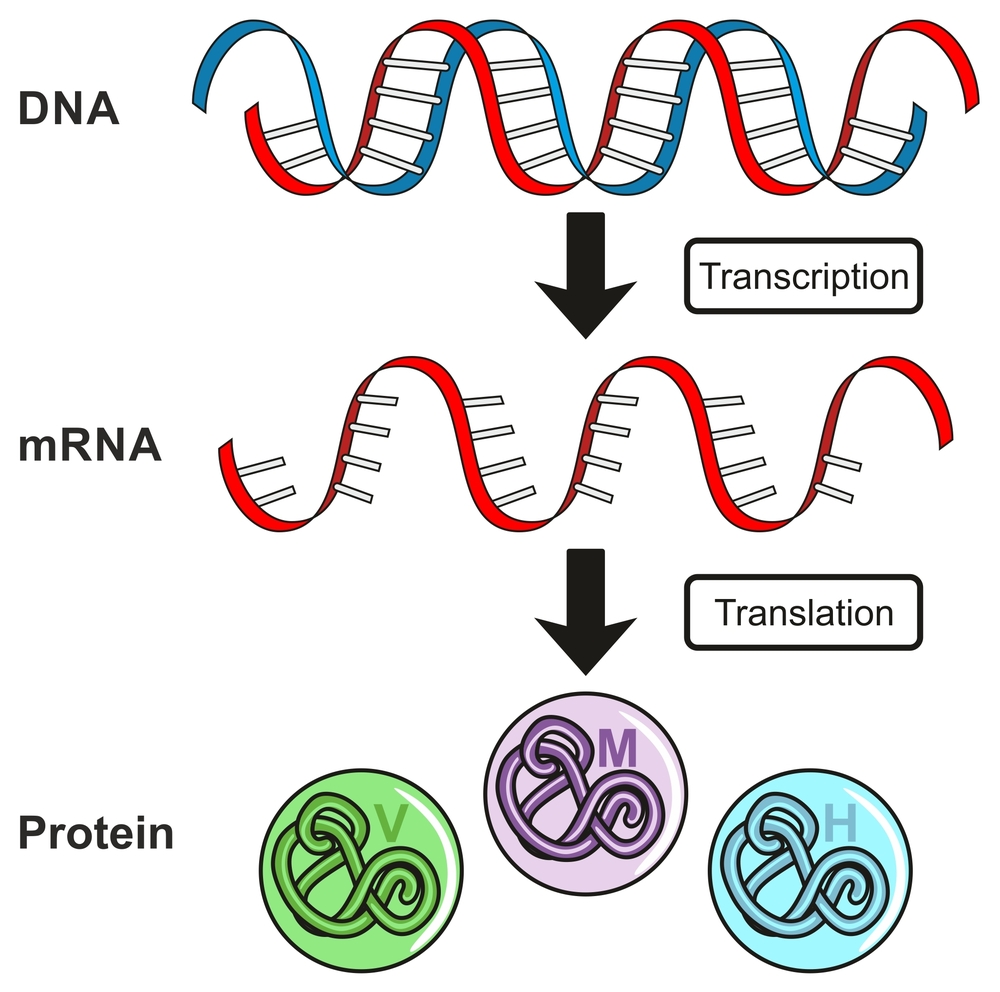
Deoxyribonucleic acid (DNA) is the elixir of life. It is like an instruction manual for protein production. The language of this manual is composed of only 4 letters: A, G, T, and C. These are the 4 nucleotide bases: adenine (A), thymine (T), guanine (G) and cytosine (C). Combinations of these 4 letters, or base pairs, in the long DNA strand, make up genes. Genes are small stretches of DNA that encode the information required for protein production via the process of transcription. These proteins are essential for the development and survival of an organism.
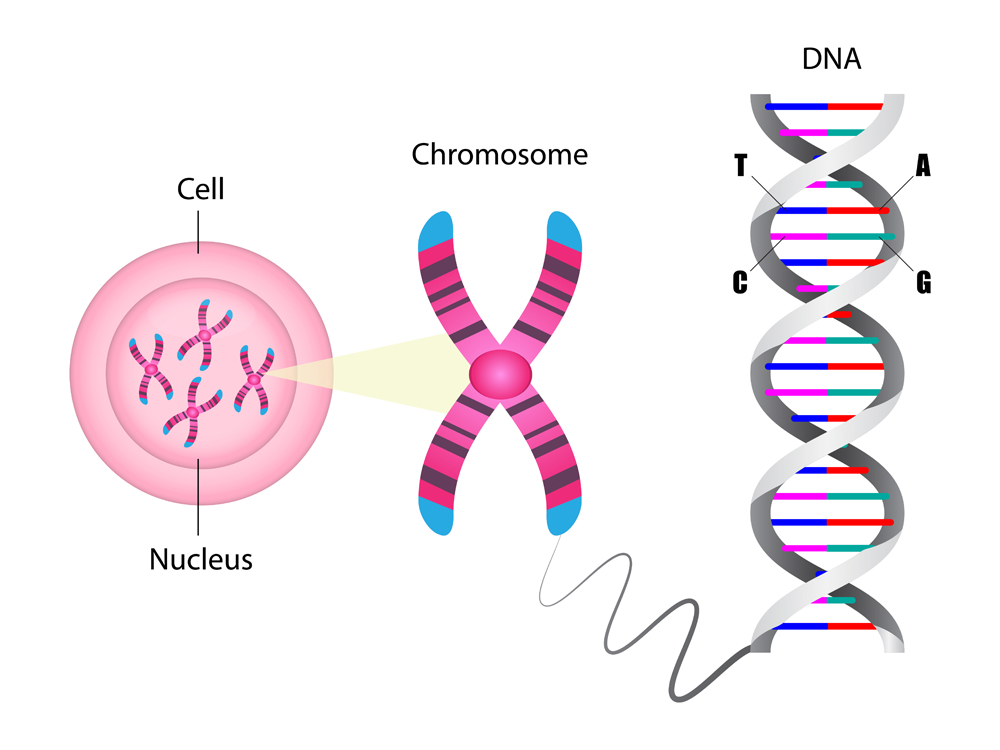
Back in 1990, several labs joined forces to decode the instructions that made up human DNA. This collaboration became known as the Human Genome Project (HGP). Upon its completion, in April 2003 we finally uncovered nature’s blueprint for constructing a human being!
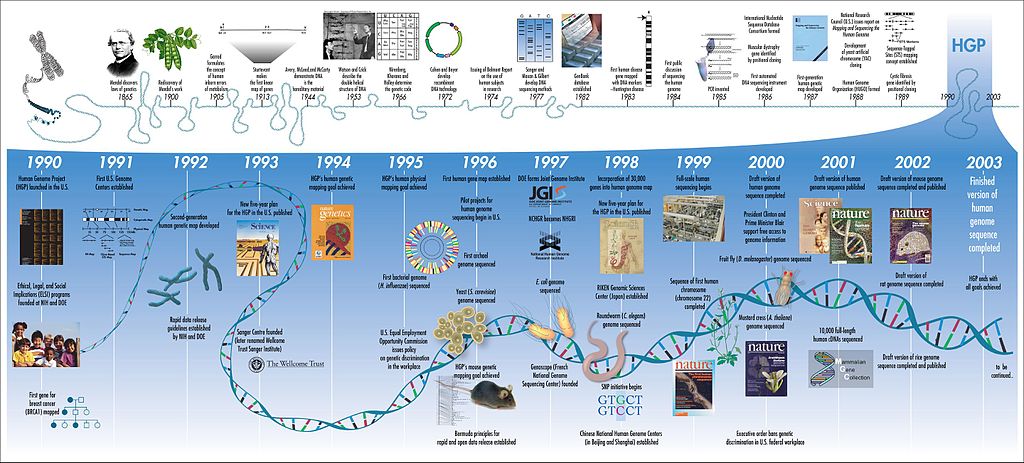
As a result of the Human Genome Project, scientists were finally able to put our DNA into numbers. The results showed that human cells comprise a whopping 3 billion base pairs of genetic material condensed into 23 pairs of chromosomes. Each of our 20,000 to 25,000 genes, which accounts for roughly 100 million base pairs, is responsible for encoding a variety of proteins that each serve a unique purpose. That’s a measly 1-2% of the entire genome coding for proteins.
The remaining 98-99% did not code for any proteins and was aptly named non-coding DNA. Since protein production was thought to be the primary role of DNA, this major chunk of DNA was also referred to as ‘junk’ DNA. However, why would the DNA—the recipe book of life—have so many pages full of gibberish?
Junk Or Undiscovered?
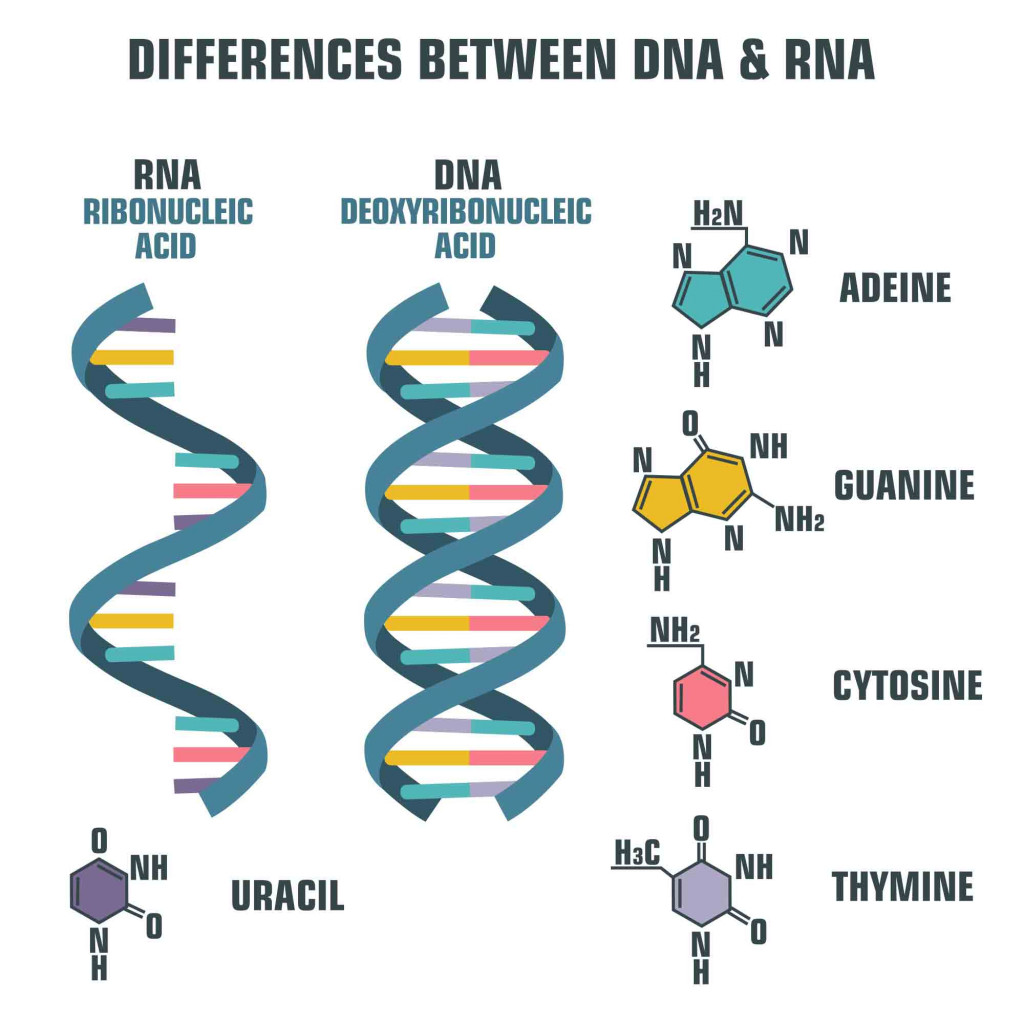
Making a protein is not as simple as following a recipe from a cookbook. Proteins are formed when DNA undergoes a process called transcription. This is required, since the enzymes that make proteins can’t read DNA. The information coded in DNA is copied onto a new molecule called messenger RNA (mRNA). Like the DNA, mRNA also has 4 nucleotide bases, but the thymine (T) is replaced by uracil (U). Another difference is that mRNA is a single-stranded molecule.
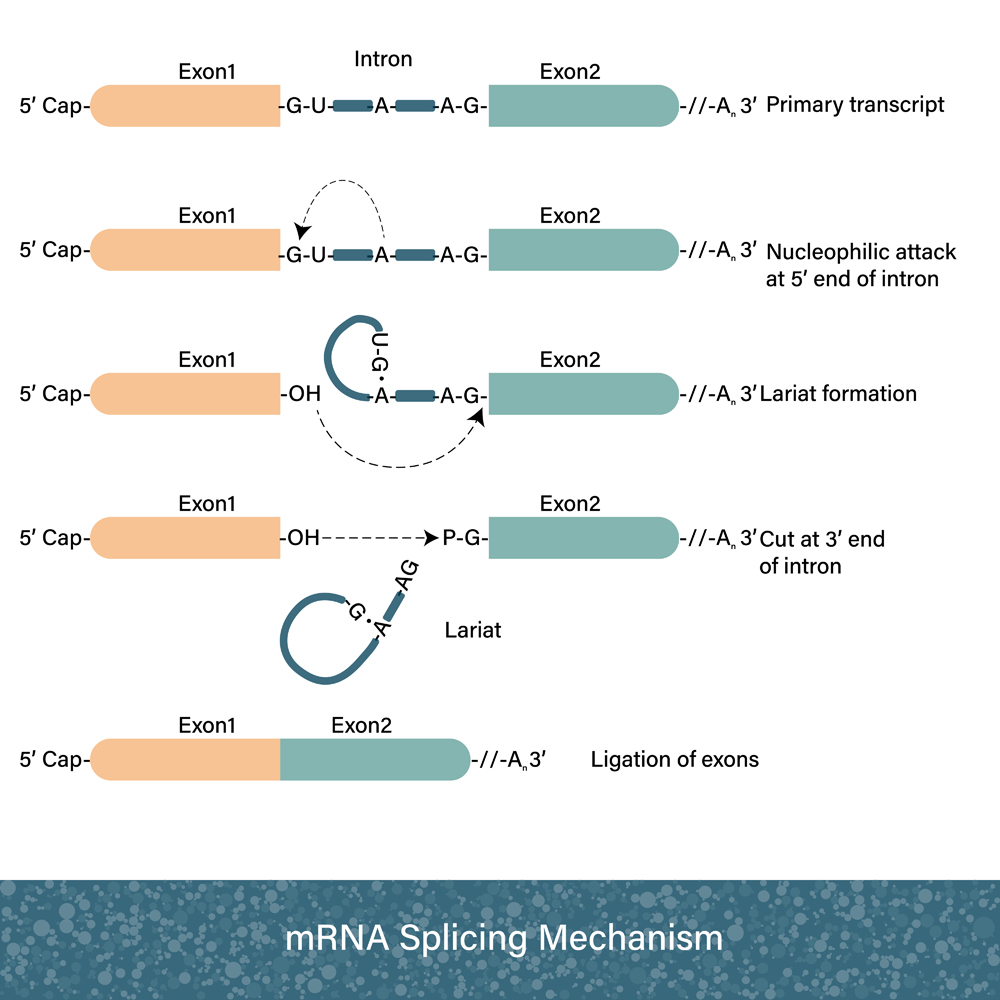
During transcription, mRNA gets chopped up and rejoined. This is known as RNA splicing. This is done because sections of the gene don’t make “protein sense”; these are called introns. During RNA splicing, these bits are cut out and discarded. You could say that these pieces are lost in transcription!
These discarded non-coding segments baffled scientists for decades. Introns were garbled nonsense between genes. Having no apparent purpose, many scientists thought it was worthless. In 1972, Susumu Ohno, a geneticist, coined the term ‘junk DNA’ to catchily explain away this DNA waste. At times, it was also called “selfish DNA”, as it seemed to exist solely for itself, without contributing anything to the organism’s survival.
However, several scientists believed that these large chunks of DNA should not be so hastily labelled as “useless”. If you are reading this article and knew only ten words of the English language, would you think that everything in this article except those ten words was nonsense? In the same way, scientists believed that the function of this so-called ‘junk’ DNA had simply yet to be discovered.

Also Read: What Does RNA Do In A Cell?
What Is The Function Of ‘Junk’ DNA?
Researchers decided to compare the human genome with the vast database of genomes of other animals. This technique is known as comparative genomics. They were shocked to find that some of the sections of junk DNA have remained unchanged, even over thousands of years. The conserved sections indicated that the non-coding DNA was essential in some way for an organism’s survival. Therefore, it was ‘positively selected’ through evolution because a mutation in this section may have proven to be detrimental.
For example, about 65 to 75 million years ago, mice and humans diverged from a common ancestor. Researchers found that out of all the conserved DNA, only 20% of it was protein-coding. The majority of the conserved DNA was actually found in non-coding portions of the genome.
However, the final nail in the coffin of junk DNA’s “uselessness” came with the Encyclopedia Of DNA Elements or ENCODE. Launched in 2003 after the completion of the Human Genome Project, ENCODE was a massive multi-lab endeavor sponsored by the National Human Genome Research Institute (NHGRI). Where the Human Genome Project sought to interpret the blueprint of human existence, ENCODE sought to figure out which sections of these blueprints really did something useful.
While the HGP used DNA sequencing to decipher the human genome, the ENCODE project looked at other elements, like RNA, through RNA sequencing, and the identification of DNA regions that may be transformed superficially by chemicals or by proteins binding to it. The project suggested that the chemical activity of a given DNA segment might provide hints about its possible function.
Recall that genes carry the information required to produce proteins, which ultimately perform cell functions. The amount of protein that a specific gene eventually produces, if any, is dictated by its gene expression. Gene expression is the ability to use the encoded information in the gene to direct protein assembly.
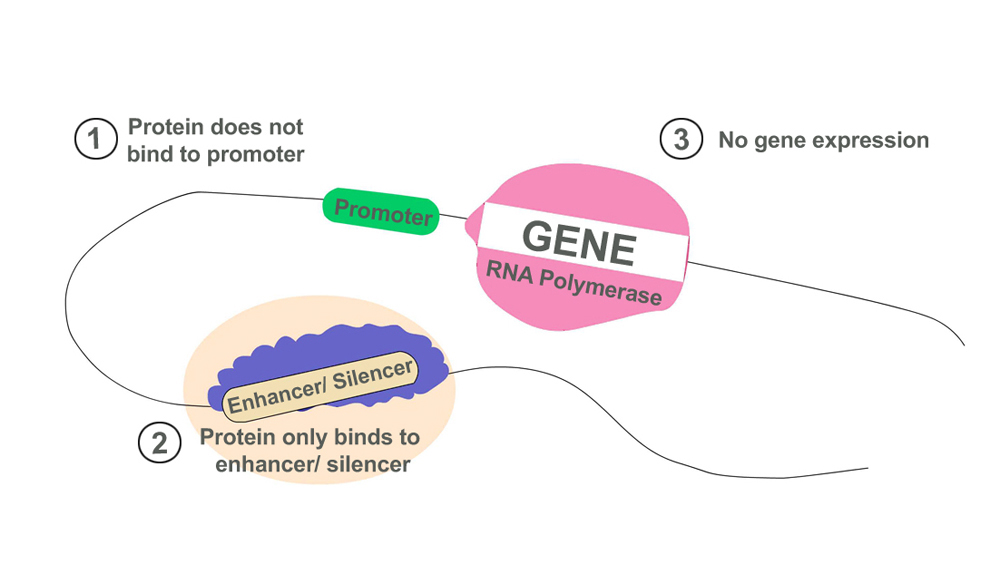
Certain proteins, transcription factors, or chemicals attach to DNA and change when and how genes are expressed. Parts of ‘junk’ DNA have been found to contain parts of DNA that regulate genes that decide when and how genes are activated and deactivated. They also act as sites for the attachment of transcription factors, which can modulate transcription. There are several types of regulatory elements that are a part of non-coding DNA, such as:
- Promoters: If a gene is a light bulb, then the promoter sequence is its switch. Promoters provide a binding site for proteins required to initiate the process of transcription. They can either switch the transcription machinery ‘on’ or ‘off’. Genes cannot produce proteins without a promoter. They are located just before the coding gene sequence.
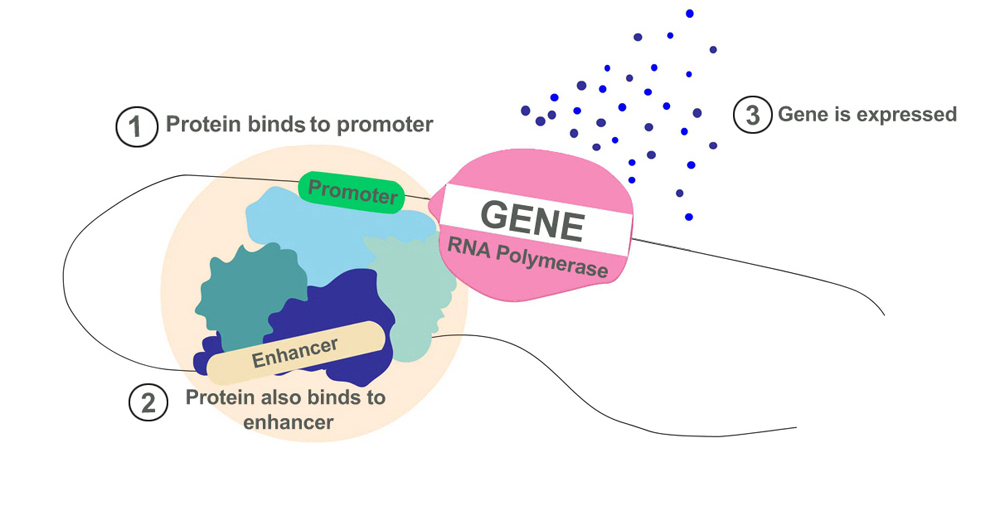
- Enhancers: Proteins that aid in the activation of transcription bind to enhancer sequences. The function of enhancers is like that of a catalyst in a chemical reaction. Transcription can still occur in the absence of an enhancer sequence, but is way more efficient in its presence. These enhancers can be found on either end of the genetic sequence or also far away.
- Silencers: Contrary to enhancers, silencers allow proteins that suppress transcription to bind to them. They prevent a gene from being over-expressed, which would result in excess proteins. They can be found at varying distances from genetic sequences, similar to enhancers.
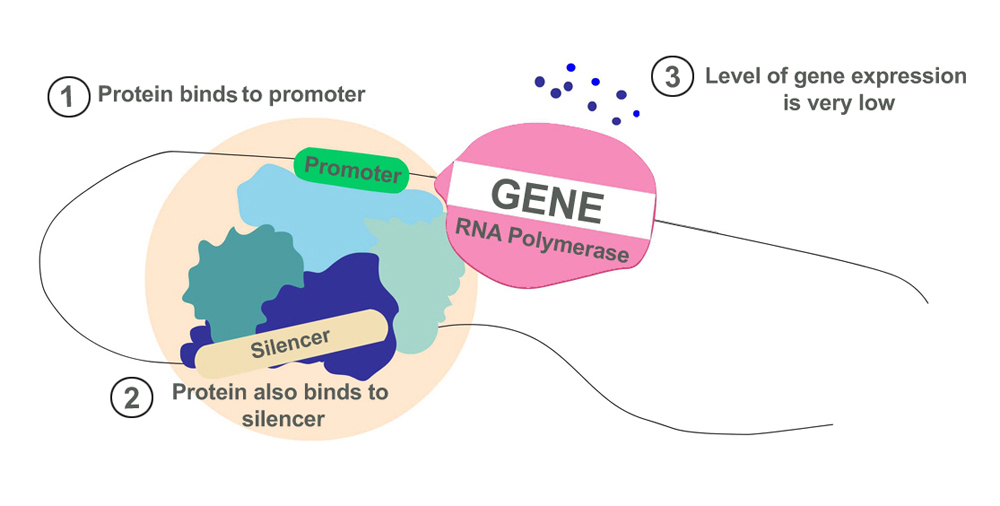
Together, enhancers and silencers function like the regulator of a fan. Instead of controlling the speed of a fan, they control the level of gene expression. The binding of proteins to enhancers is like turning the knob to full speed, whereas binding to silencers brings the ‘fan’ to a halt.
These are just a few examples of the ‘functions’ of non-coding DNA. So, is a non-coding DNA a functional DNA sequence?
A ‘functional’ DNA sequence is one that can control gene expression, that is, the amount of protein produced by a particular gene sequence. It is this variation in protein composition that makes each cell type unique. Therefore, as each cell consists of the same genome and DNA, it is the gene expression levels that decide if a cell is a skin cell, immune cell, neuron, etc.
The ENCODE consortium carried out these techniques in a variety of cell types to account for the inherent variability. So, in accordance with this definition of ‘functional’, ‘junk’ DNA definitely plays a role in influencing gene expression. The results of the ENCODE project revealed just how little we knew about the mysterious non-coding section of the genome.
In 2012, the efforts of the ENCODE consortium revealed that over 80% of the genomic bases showcased biochemical activity. Hence, there was certainly a biological function for far more than the 1% of our DNA that constitutes genes. The project has revealed a remarkable collection of previously unidentified signals and switches embedded like tattoos over the complete length of the human DNA.
Also Read: Why Was The Human Genome Mapping Project Stuck At 92% For So Many Years?
Conclusion
Since the ENCODE consortium has published their results, scientists have associated non-coding DNA sequences to different biological processes and human diseases. Researchers hypothesize that these sequences underlie the development of our opposing thumbs, as well as the uterus! A paper published in Oncogene has also shown that a non-coding segment of DNA regulates gene expression, eventually impacting the risk of prostate and breast cancer. Hence, uncovering the full function of so-called junk DNA has become an intensive field of research.
However, it is important to note that ENCODE’s meaning of “functional” is hotly debated. Many scientists say that the results of the ENCODE project are misleading and vastly overestimated. They state that a protein merely binding to DNA or undergoing a chemical change does not guarantee that the sequence serves a meaningful role. In the body, several DNA-protein binding events are often random and insignificant, casting a shadow of doubt over the results published by the consortium.
Keeping these justified criticisms in mind, it will take many more research studies to quantify the functional capabilities of non-coding DNA. However, there is no denying the fact that ‘junk’ DNA is not really junk at all!
How well do you understand the article above!

References (click to expand)
- How so-called 'junk DNA' affects cancer risk | News. The Harvard T.H. Chan School of Public Health
- What is noncoding DNA? - Genetics - MedlinePlus. MedlinePlus
- The 99 Percent… of the Human Genome - Science in the News. Harvard University
- The Human Genome Project. The National Human Genome Research Institute
- CASTILLODAVIS, C. (2005, October). The evolution of noncoding DNA: how much junk, how much func?. Trends in Genetics. Elsevier BV.
- ENCODE-funded Publications - UCSC Genome Browser. University of California, Santa Cruz
- Introduction - Mapping and Sequencing the Human Genome. The National Center for Biotechnology Information
- The ENCODE Project Consortium. (2012, September). An integrated encyclopedia of DNA elements in the human genome. Nature. Springer Science and Business Media LLC.
